StrainTool allows the estimation of Strain Tensor parameters, on the earth's crust, given a list of data point, aka points on the earth along with their tectonic velocities.
StrainTool has three basic components:
pystrainStrainTensor.pyStrainTensor.pypystrain is a package written in python,
defining a variety of functions, classes, etc to enable the computation of
strain tensors.
When installing this package, all of its modules will be available to
the user. It is the core part of the project and should be installed
before the main program (StrainTensor.py) can be used.
The package is found under StrainTool/pystrain
This is the main program, i.e the program that the user will
run to produce strain tensor results. It is heavily dependant on the
pystrain package (so any change there will affect
StrainTensor.py's behaviour
This program does not need installation; it is just a pyrhon script. The user may only need to change priviledges to make it executable
StrainTensor.py is located at StrainTool/bin
By 'Plot Tools', we mean an array of shell (Bash)
scripts that can be used to plot StrainTensor.py results.
These scripts actualy use GMT to
perform the plotting. The scripts are an efficient, ready-to-use
It is by no means mandatory to use these scripts to plot the results; users can use their own tools and/or scripts to do so. Actualy, this is a totaly independent part of the Project and can be skipped alltogether.
These scripts, are found under StrainTool/plot
To install the Project, you will need:
NumPy and SciPy have extended installation guides
for most, if not all, operating systems.
To use the Plot Tools bundle, you must have GMT (> 5.0.0) installed, allong with a variety of UNIX-based tools.
To install the Project, you actualy only need to install the
pystrain package. What this means, is that
python will see that the package modules are build and extracted to a suitable folder
on your local system (something like /usr/lib/python2.7/site-packages/ on
a UNIX-like system). After this step, you will be able to make use of the
modules in the package, both for running StrainTool.py and
for any new program you may wish to code. StrainTensor.py
and Plot Tools are programs, so you do not
have to "install" them.
To install pystrain, do the following:
StrainTool/pystrainsetup.py script, so you only need
to run it (as root/administrator): $> python setup.py install$> python -c "import pystrain"The following scenarios have been tested to validatethe installation procedure
| OS | Python 2.7 | Python 3.6 | GMT 5.2 | GMT 5.4 |
|---|---|---|---|---|
| Fedora | ✔ | ✔ | ||
| Manjaro | ✔ | ✔ | ||
| Ubuntu | ✔ | ✔ | ✔ | ✔ |
| Windows 10 | ✔ | ✔ | ✔ | ✔ |
To validate that the installation has succeded, you can run a "test case"
scenario. Under the folder data/, you will find a file named
CNRS_midas.vel. This ascii file, contains velocity and coordinate information
for a large list of GPS/GNSS stations around the globe. We will use it to
perform a test run of StrainTensor.py.
Go to the bin/ folder and execute the command:
$> ./StrainTensor.py -i ../data/CNRS_midas.vel -r 18.75/30.25/32.75/42.25 --x-grid-step=0.5 --y-grid-step=0.5 --dmin=1 --dmax=500 --dstep=1 --Wt=24 -c -g CNRS_midas.vel.
The results will be written in a (newly created) file, named strain_info.dat,
while station information will be written in the file station_info.dat
(this latter file is mainly used for plotting). You should be able to verify that
both files have been succesefuly created.
Under data/ you will find the "reference" output files for checking,
named strain_info.dat.ref and station_info.dat.ref.
Verify that the files you have just created (placed under bin/)
contain the same results as the "reference" files (this can be very easily
performed using the diff command).
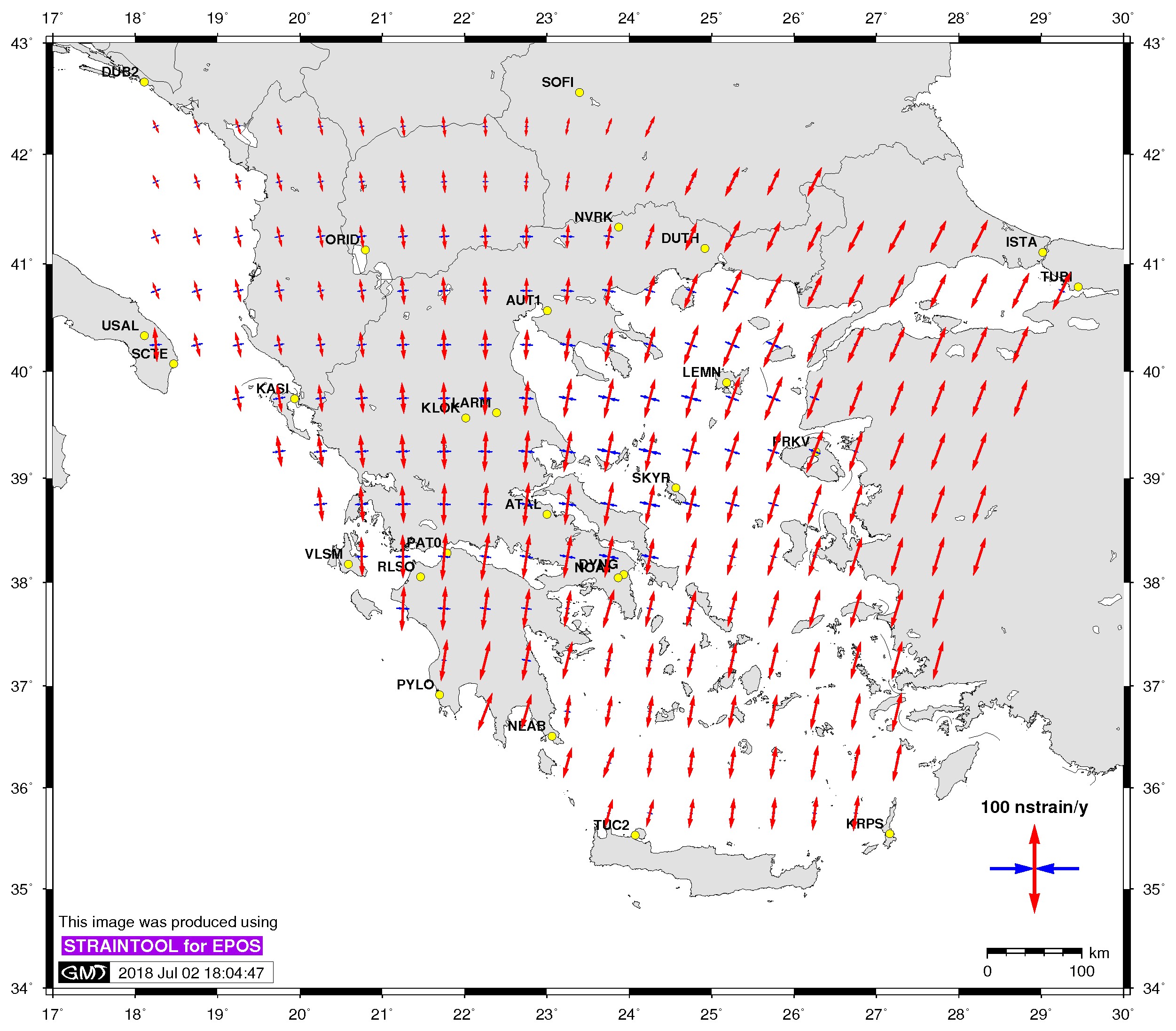
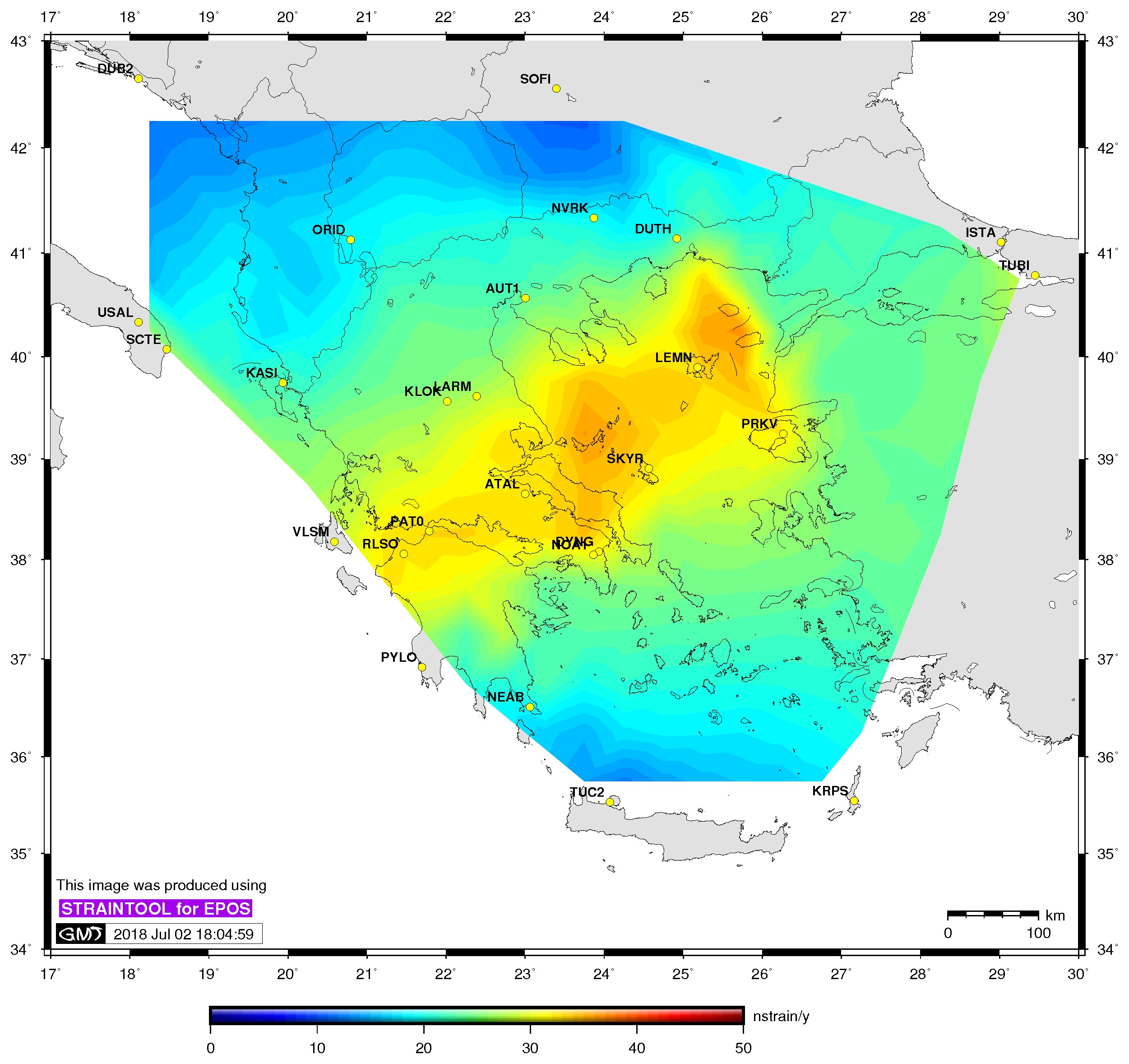
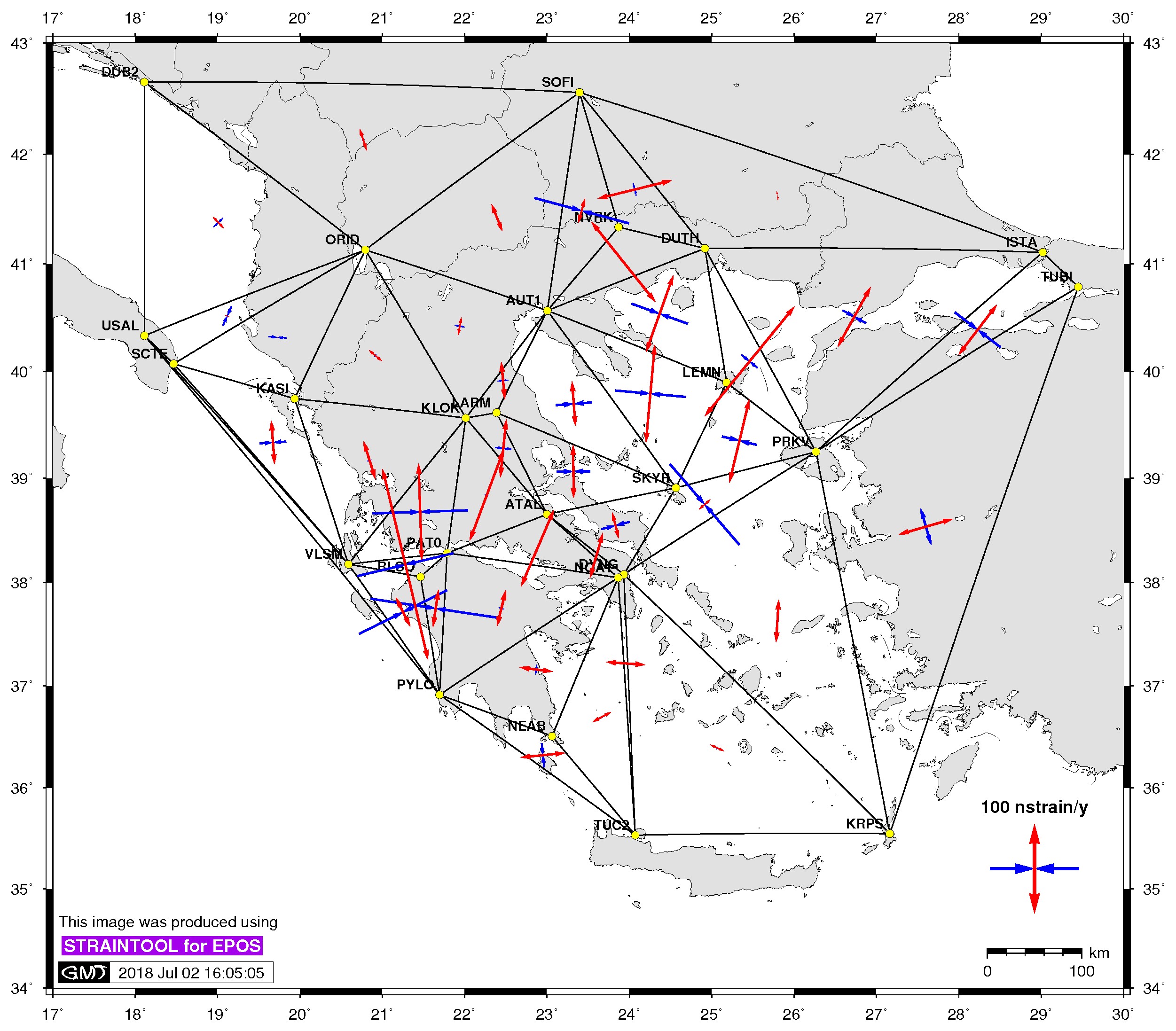
| Principal axis | Shear strain | Delaunay triangles |
|---|---|---|
$> ./gmtstrainplot.sh -jpg -str strain_info.dat -psta -l -o output_str |
$> ./gmtstrainplot.sh -jpg -gtot strain_info.dat -psta -l -o output_gtot |
$> ./gmtstrainplot.sh -jpg -str strain_info.dat -psta -l -o output_str -deltr |
 |
 |
 |
StrainTensor.py computes strain tensor parameters on a selected region. The
program comes with an extended help message, that can be displayed via the
-h or --help switch (aka
$> ./StrainTensor.py [...] --helpStrainTensor.py's
behaviour via a variety of available switches.
To use the program, the only mandatory switch is the -i or --input-file, which specifies the input file to be used. All other options will automaticaly assume default values (see Options for a detailed list). Basic usage is described below:
usage: StrainTensor.py [-h] -i INPUT_FILE [--x-grid-step X_GRID_STEP]
[--y-grid-step Y_GRID_STEP] [-m METHOD] [-r REGION]
[-c] [-b] [--max-beta-angle MAX_BETA_ANGLE]
[-t WEIGHTING_FUNCTION] [--Wt Wt] [--dmin D_MIN]
[--dmax D_MAX] [--dstep D_STEP] [--d-param D_PARAMETER]
[-g] [--verbose] [--multicore] [-v]
The whole list of available options, is:
optional arguments:
-h, --help show this help message and exit
-i INPUT_FILE, --input-file INPUT_FILE
The input file. This must be an ascii file containing the columns: 'station-name longtitude latitude Ve Vn SigmaVe SigmaVn Sne time-span'. Longtitude and latitude must be given in decimal degrees; velocities (in east and north components) in mm/yr. Columns should be seperated by whitespaces. Note that at his point the last two columns (aka Sne and time-span) are not used, so they could have random values.
--x-grid-step X_GRID_STEP
The x-axis grid step size in degrees. This option is only relevant if the program computes more than one strain tensors. Default is 0.5(deg).
--y-grid-step Y_GRID_STEP
The y-axis grid step size in degrees. This option is only relevant if the program computes more than one strain tensors. Default is 0.5(deg)
-m METHOD, --method METHOD
Choose a method for strain estimation. If 'shen' is passed in, the estimation will follow the algorithm described in Shen et al, 2015, using a weighted least squares approach. If 'veis' is passed in, then the region is going to be split into delaneuy triangles and a strain estimated in each barycenter. Default is 'shen'.
-r REGION, --region REGION
Specify a region; any station (in the input file) falling outside will be ommited. The region should be given as a rectangle, specifying min/max values in longtitude and latitude (using decimal degrees). E.g. "[...] --region=21.0/23.5/36.0/38.5 [...]"
-c, --cut-excess-stations
This option is only considered if the '-r' option is set. If this this option is enabled, then any station (from the input file) outside the region limit (passed in via the '-r' option) is not considered in the strain estimation.
-b, --barycenter Only estimate one strain tensor, at the region's barycentre.
--max-beta-angle MAX_BETA_ANGLE
Only relevant for '--mehod=shen'. Before estimating a tensor, the angles between consecutive points are computed. If the max angle is larger than max_beta_angle (in degrees), then the point is ommited (aka no tensor is computed). This option is used to exclude points from the computation tha only have limited geometric coverage (e.g. the edges of the grid). Default is 180 deg.
-t WEIGHTING_FUNCTION, --weighting-function WEIGHTING_FUNCTION
Only relevant for '--mehod=shen'. Choose between a 'gaussian' or a 'quadratic' spatial weighting function. Default is 'gaussian'.
--Wt Wt Only relevant for '--mehod=shen' and if 'd-param' is not passed in. Let W=Σ_i*G_i, the total reweighting coefficients of the data, and let Wt be the threshold of W. For a given Wt, the smoothing constant D is determined by Wd=Wt . It should be noted that W is a function of the interpolation coordinate, therefore for the same Wt assigned, D varies spatially based on the in situ data strength; that is, the denser the local data array is, the smaller is D, and vice versa. Default is Wt=24.
--dmin D_MIN Only relevant for '--mehod=shen' and if 'd-param' is not passed in. This is the lower limit for searching for an optimal d-param value. Unit is km. Default is dmin=1km.
--dmax D_MAX Only relevant for '--mehod=shen' and if 'd-param' is not passed in. This is the upper limit for searching for an optimal d-param value. Unit is km. Default is dmax=500km.
--dstep D_STEP Only relevant for '--mehod=shen' and if 'd-param' is not passed in. This is the step size for searching for an optimal d-param value. Unit is km. Default is dstep=2km.
--d-param D_PARAMETER
Only relevant for '--mehod=shen'. This is the 'D' parameter for computing the spatial weights. If this option is used, then the parameters: dmin, dmax, dstep and Wt are not used.
-g, --generate-statistics
Only relevant when '--mehod=shen' and '--barycenter' is not set. This option will create an output file, named 'strain_stats.dat', where estimation info and statistics will be written.
--verbose Run in verbose mode (show debugging messages)
--multicore Run in multithreading mode (default: False)
-v Display version and exit.
For example, the command we used on the Example section:
$> ./StrainTensor.py -i ../data/CNRS_midas.vel -r 18.75/30.25/32.75/42.25 --x-grid-step=0.5 --y-grid-step=0.5 --dmin=1 --dmax=500 --dstep=1 --Wt=24 -cTo perform the computations, StrainTensor.py needs an input file,
that holds input data. Usually, this implies a list of GPS/GNSS stations with
their ellipsoidal coordinates (aka longtitude and latitude) and their respective
tectonic velocities (usually estimated using position time-series) allong with
the corresponding standard deviation values. The format of these files,
should follow the convention:
station-name longtitude latitude Ve Vn SigmaVe SigmaVn Sne time-span
string deg. deg. mm/yr mm/yr mm/yr mm/yr / dec. years
Station coordinates are provided in longtitude/latitude pairs in decimal degrees.
Velocities and velocity standard deviations are provided in mm per years (mm/yr).
Sne is the correlation coefficient between East and North velocity components
and time-span is the total time span of the station timeseries in decimal
degrees. Sne
and time-span) are not used, so they could have random values.There are no strict formating rules on how the individual elements should be
printed (i.e. how many fields, decimal places, etc). The only condition is
that fields are seperated by whitespace(s). To see an example of a valid
input file, you can check data/CNRS_midas.vel.
Results of StrainTensor.py are recorded in the following three files:
Latitude Longtitude vx+dvx vy+dvy w+dw exx+dexx exy+dexy eyy+deyy emax+demax emin+demin shr+dshr azi+dazi dilat+ddilat sec.inv+dsec.inv deg deg mm/yr mm/yr deg/Myr nstrain/yr nstrain/yr nstrain/yr nstrain/yr nstrain/yr nstrain/yr deg. nstrain/yr nstrain/yr
Code Longtitude Latitude Ve Vn dVe dVn string deg deg mm/yr
Plotting scripts are placed under plot/ directory. They are:
gmtstrainplot.sh,gmtstatsplot.sh, andplotall.shdefault-param to be in the
same folder.
default-param file options
pth2inptf=../data/ # set default folder for input files (strain_info.dat, station_info.dat)
west, east, south, north, projscale, frame, sclength: set region parameters
STRSC=0.01 : set principal axis plot scale
ROTSC=.7 : set rotational rates plot scale
gmtstrainplot.sh options:
Basic Plots & Background :
-r | --region : region to plot (default Greece)
usage: -r west east south north projscale frame
PLot station and velocitiess:
-psta [:=stations] plot only stations from input file
-vhor (station_file)[:= horizontal velocities]
-vsc [:=velocity scale] change valocity scale default 0.05
Plot strain tensor parameters:
-str (strain file)[:= strains] Plot strain rates
-rot (strain file)[:= rots] Plot rotational rates
-gtot(strain file)[:=shear strain] plot total shear strain rate contours
-gtotaxes (strain file) dextral and sinistral max shear strain rates
-dil (strainfile)[:= dilatation] Plot dilatation and principal axes
-secinv (strain file) [:=2nd invariand] Plot second invariand
-strsc [:=strain scale]
-rotsc [:=rotational scales]
*for -gtot | -dil | -secinv use +grd to plot gridded data
ex:-gtot+grd
Other options:
-o | --output : name of output files
-l | --labels : plot labels
-mt | --map_title "title" : title map default none, use quotes
-jpg : convert eps file to jpg
-h | --help : help menu
For example, to plot the principal axis fo strain rates for the example case above you can use the following command:
$>./gmtstrainplot.sh -jpg -str strain_info.dat -psta -l
gmtstatsplot.sh options:
Basic Plots & Background :
-r | --region : region to plot (default Greece)
usage: -r west east south north projscale frame
Plot Stations and triangles:
-psta [:=stations] plot only stations from input file
-deltr [:= delaunay triangles] plot delaunay triangles
Plot Statistics:
-stats (input file) set input file
--stats-stations : plot used stations
--stats-doptimal : plot optimal distance (D)
--stats-sigma : plot sigma
Other options:
-o | --output : name of output files
-l | --labels : plot labels
-leg : plot legends
-mt | --map_title : title map default none use quotes
-jpg : convert eps file to jpg
-h | --help : help menu
For example, to plot the stations used per cell grid for the example case above you can use the following command:
$> ./gmtstatsplot.sh -jpg -stats strain_stats.dat --stats-stations -leg -o output_stats-stations
plotall.sh options:
- no switch produce default output names
-p | --prefix : add prefix work id to output file
-s | --suffix : add suffix work id to output file
-h | --help: help panel
For example:
$> ./plotall.sh -p test -s test2
Given a set of stations (aka point on earth's surface) with their corresponding east and north velocities, we can estimate (or compute) strain tensor parameters, by solving for the system $$ \begin{bmatrix} V_{x,S_1} \\ V_{y,S_1} \\ V_{x,S_2} \\ V_{y,S_2} \\ \cdots \\ V_{x,S_n} \\ V_{y,S_n} \\ \end{bmatrix} = \begin{bmatrix} 1 & 0 & \Delta_{y_1} & \Delta_{x_1} & \Delta_{y_1} & 0 \\ 0 & 1 & -\Delta_{x_1} & 0 & \Delta_{x_1} & \Delta_{y_1} \\ 1 & 0 & \Delta_{y_2} & \Delta_{x_2} & \Delta_{y_2} & 0 \\ 0 & 1 & -\Delta_{x_2} & 0 & \Delta_{x_2} & \Delta_{y_2} \\ \cdots & \cdots & \cdots & \cdots & \cdots & \cdots \\ 1 & 0 & \Delta_{y_n} & \Delta_{x_n} & \Delta_{y_n} & 0 \\ 0 & 1 & -\Delta_{x_n} & 0 & \Delta_{x_n} & \Delta_{y_n} \\ \end{bmatrix} \begin{bmatrix} U_{x} \\ U_{y} \\ \omega \\ \tau_{x} \\ \tau_{xy} \\ \tau_{y} \\ \end{bmatrix} $$ at any given location \(R\); \(\Delta_{x_i}\) and \(\Delta_{y_i}\) are the displacement components between station \(i\) and the point \(R\). A minimum of 3 stations is required to compute the parameters; if more than 3 stations are used, then the parameters are estimated (here, using a least squares approach).
Assuming that we have variance information for the station velocities (and a Gaussian distribution), we can add the covariance matrix \(C\) of the velocity data in the system. In the simplest case, \(C\) is a diagonal matrix, with the velocity component standard deviations as its elements, $$ C = {\sigma_{0}}^2 \begin{bmatrix} (1/\sigma_{V_{x,S_{1}}})^2 & 0 & 0 & \cdots & 0 \\ 0 & (1/\sigma_{V_{y,S_{1}}})^2 & 0 & \cdots & 0 \\ 0 & 0 & (1/\sigma_{V_{x,S_{2}}})^2 & \cdots & 0 \\ \vdots & \vdots & \vdots & \ddots & \vdots \\ 0 & 0 & 0 & \cdots & (1/\sigma_{V_{y,S_{n}}})^2 \\ \end{bmatrix} $$
Shen et al, 2015, propose a more elaborate approach, reconstructing the covariance matrix by multiplying a weighting function to each of its diagonal terms. The weighting function \(G_i = L_i \cdot Z_i\), in which \(L_i\) and \(Z_i\) are functions of distance and spatial coverage dependent, respectively. The final covariance matrix becomes then, \(C = C \cdot G^{-1}\) or, since its diagonal, \(C_i = C_i \cdot {G_i}^{-1}\)
The following paragraphs, describe the estimation process when using shen method (i.e. --method=shen) as the algorithm of choice.
In coarse lines, when choosing the shen method, the program will
'girddify' the region of choise and estimate one strain tensor at the centre of each cell.
The region of choise can be given via the --region switch, using
minimum and maximum longtitude and latitude as decimal degrees, using '/' as
delimeter. E.g. [...] --region=21.0/23.5/36.0/38.5 [...]. If the region
is not specified, then the maximum and minimum longtitude values will be extracted from the
list of input stations, aka the region will be defined to include all stations
within the input file.
The user can specify the grid step size, both in x and y components, via the --x-grid-step and --y-grid-step switches respectively. The step sizes should be given in decimal degrees and if not set, have a default value of \(0.5 deg.\)
The program will try to estimate strain tensor parameters at the centre of each cell. The algorithm depends on a number of options that the user can define via the command line arguments. Using a spatial smoothing coefficient \(D\), either defined by the user (--d-param) or found by the program (see Finding the optimal smoothing parameter \(D\)), the covariance matrix will be formed, using distance-dependent weighting \(L_i\), spatial weighting \(Z_i\) and the data variance information \(C_i\). The final covariance matrix, will be \(C \cdot G^{-1}\) where \(G_i = L_i \cdot Z_i\).
Strain tensor parameters will be estimated using a (weighted) least squares approach;
Six fundamental parameters are solved for and in a next step values of interest
are computed (see Strain Tensor Parameters).
Computed values are written to the output file strain_info.dat (see Output Files).
For distance dependent weighting \(L_i\), we can use two formulas, $$L_i = \exp(-{\Delta}R_i^{2} / D^{2})$$ or $$L_i = 1/(1 + {\Delta}R_i^{2} / D^{2})$$ n which a spatial smoothing parameter \(D\) is introduced (see Finding the optimal smoothing parameter). According to Shen et al, 2015,
Both functions allow reduced weight of the data as distance increases. The difference between the two is that the Gaussian function reduces the weight at a faster pace with distance \(\Delta R_i\) than that of the quadratic function. Depending on the data quality, the Gaussian function can offer a relatively finer resolution of the interpolation result if the data are clean and smooth. On the other hand, if the data are somewhat heterogeneous, the quadratic function is relatively more conservative and provides a more smoothed solution, especially for regions in which data are sparsely distributed.
To actually compute the distance-dependent weights \(L_i\), we need the smoothing coefficient \(D\). Users have two options,
Any data points with distance exceeding a threshold value \(L_{max}\) are excluded from the estimation. The value of \(L_{max}\) depends on the Distance-dependent weighting function; if a Gaussian approach is selected, then \(L_{max}=2.15 \cdot D km \), while for the quadratic approach \(L_{max}=10 \cdot D km \) , where \(D\) is the spatial smoothing parameter.
The spatial weighting function measures the azimuth span of the site with respect to each of the data points. The azimuth span \(\theta\) for data point \(i\) is measured between two strike directions of the \(i−1\) and \(i+1\) data points in its neighborhood.
When using --method=veis a different aproach is followed. The region is split into Delaunay triangles at the barycentre of which a strain tensor is computed. Note that this approach only uses three points (aka the triangle points) to calculate tensor parameters, so the parameters are computed and not estimated. Covariance info is not used at all with this approach.
Note that when using this approach,
strain_info.dat file (remember this a computation not an estimation),delaunay_info.dat,TODO
The computation/estimation process, solves for the vector of parameters \([U_{x}, U_{y}, \omega, \tau_{x}, \tau_{xy}, \tau_{y}]^T\). The rest parameters of interest, written in the output file , are computed as follows: $$ \tau_{max}= \sqrt{ {\tau_{xy}}^2 + {e_{diff}}^2 } $$ $$ e_{max} = e_{mean} + \tau_{max} $$ $$ e_{min} = e_{mean} - \tau_{max} $$ $$ Az_{e_{max}} = 90 + \frac{-atan2(\tau_{xy}, e_{diff})}{2} $$ $$ dilatation = \tau_{x} + \tau_{y} $$ $$ second invariant = \sqrt{ {\tau_{x}}^2 + {\tau_{y}}^2 + 2{\tau_{xy}}^2 }$$ where, $$ e_{mean} = \frac{\tau_{x}+\tau_{y}}{2} $$ $$ e_{diff} = \frac{\tau_{x}-\tau_{y}}{2} $$
git checkout -b my-new-idea)git commit -am 'Add some feature')git push origin my-new-idea)The work is licensed under MIT-license
Dr. Dimitrios G. Anastasiou
Dr. Rural & Surveying Engineer | Dionysos Satellite Observatory - NTUA | dganastasiou@gmail.com
Xanthos Papanikolaou
Rural & Surveying Engineer | Dionysos Satellite Observatory - NTUA | xanthos@mail.ntua.gr
Dr. Athanassios Ganas
Research Director | Institute of Geodynamics | National Observatory of Athens | aganas@gein.noa.gr
Prof. Demitris Paradissis
Professor NTUA | Dionysos Satellite Observatory - NTUA | dempar@central.ntua.gr
The history of releases can be viewed at ChangeLog
EPOS IP - EPOS Implementation Phase
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement N° 676564
Disclaimer: the content of this website reflects only the author’s view and the Commission is not responsible for any use that may be made of the information it contains.
Anastasiou D., Ganas A., Legrand J., Bruyninx C., Papanikolaou X., Tsironi V. and Kapetanidis V. (2019). Tectonic strain distribution over Europe from EPN data. EGU General Assembly 2019, Geophysical Research Abstracts, Vol. 21, EGU2019-17744-1 Abstract
Shen, Z.-K., M. Wang, Y. Zeng, and F. Wang, (2015), Strain determination using spatially discrete geodetic data, Bull. Seismol. Soc. Am., 105(4), 2117-2127, doi: 10.1785/0120140247
Veis, G., Billiris, H., Nakos, B., and Paradissis, D. (1992), Tectonic strain in greece from geodetic measurements, C.R.Acad.Sci.Athens, 67:129--166
Python Software Foundation. Python Language Reference, version 2.7. Available at http://www.python.org